Software
Reproducible Research
I not only hope that this page can be useful to you but it also may encourage you to make your research accessible and reproducible. I followed the guideline to reproducible research by Patrick Vandewalle.
Quick links to all software
- Multi-Contrast MRI Reconstruction with Structure-Guided Total Variation
- Joint reconstruction of PET-MRI by exploiting structural similarity
- Vector-Valued Image Processing by Parallel Level Sets
- Blind Image Fusion for Hyperspectral Imaging with the Directional Total Variation
- Stochastic PDHG with Arbitrary Sampling and Imaging Applications
- Enhancing the spatial resolution of hyperpolarized carbon‐13 MRI of human brain metabolism using structure guidance
Multi-Contrast MRI Reconstruction with Structure-Guided Total Variation
Download software: [7 MB]
Tested Configurations: The software was tested on Ubuntu 12.04 LTS with Matlab 2015a and Mac OS X with Matlab 2015a.
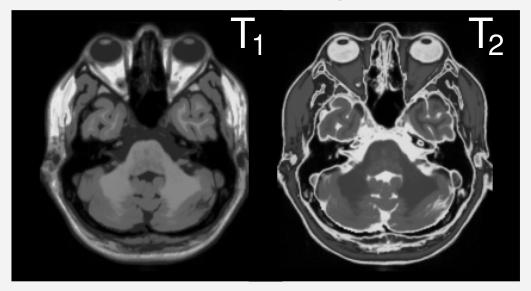
Motivation: Different scan parameters lead to different contrasts in MRI.
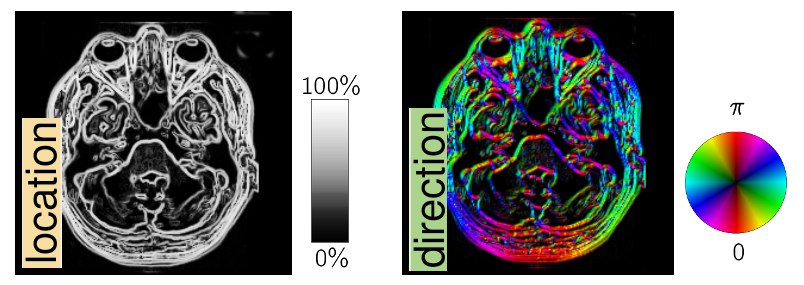
Idea: Structure can be either based on the location or direction of edges.
Acknowledgements: The simulation results are based on BrainWeb data and patient data kindly provided by Ninon Burgos and Jonathan Schott from the University College London, UK.
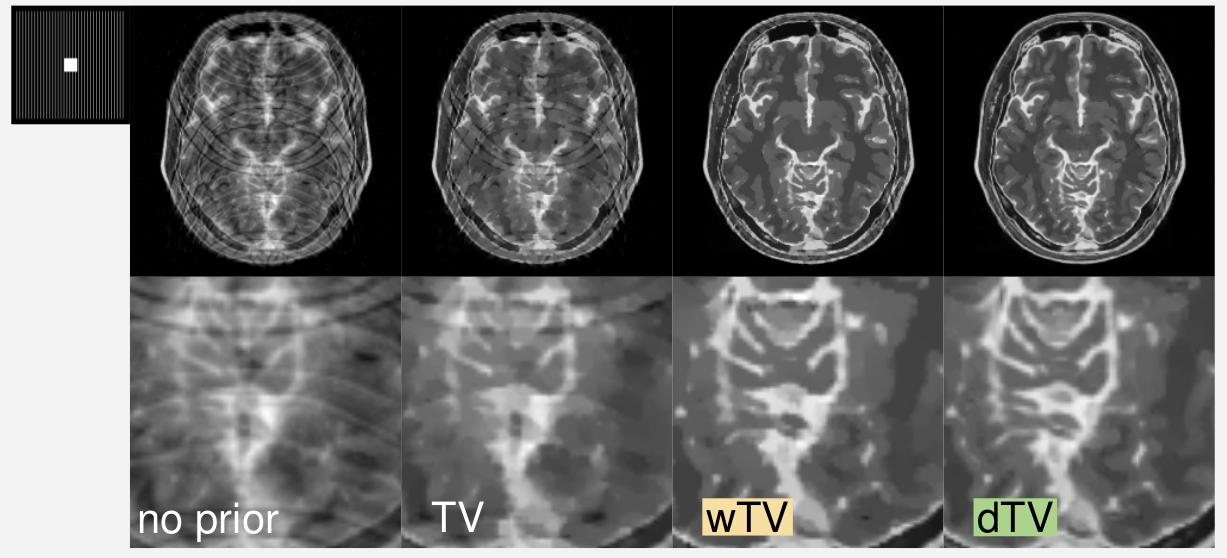
Example Result: Reconstructions with different a priori knowledge. The reconstruction quality improves visually significantly from left to right.
Joint Reconstruction of PET-MRI by Exploiting Structural Similarity
Paper: [IOP science]
Download software: [link, 59 MB]
Tested Configurations: The software was tested on Ubuntu 12.04 LTS with Matlab 2013a.
Reference: M. J. Ehrhardt, K. Thielemans, L. Pizarro, D. Atkinson, S. Ourselin, B. F. Hutton and S. R. Arridge, Joint reconstruction of PET-MRI by exploiting structural similarity, Inverse Problems 31(1), 015001, 2015
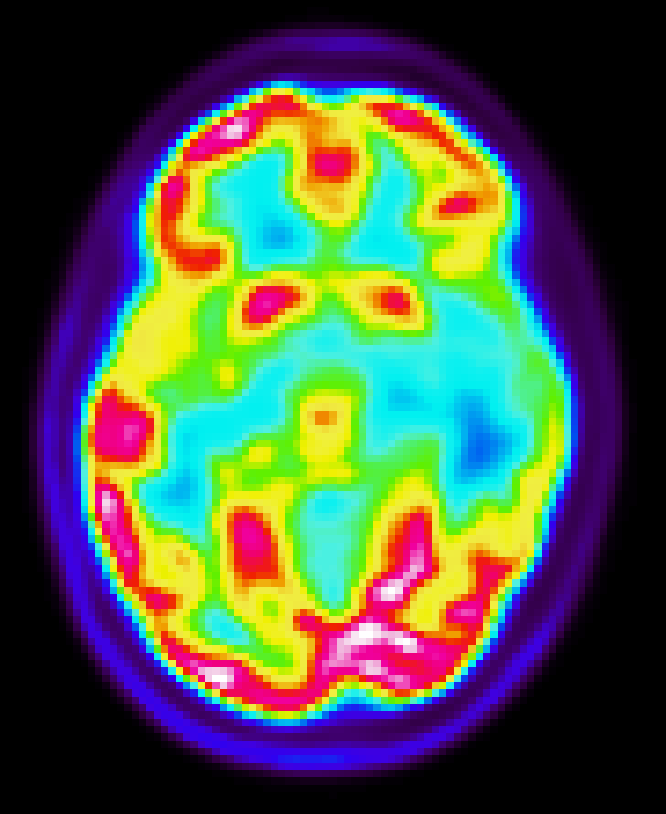
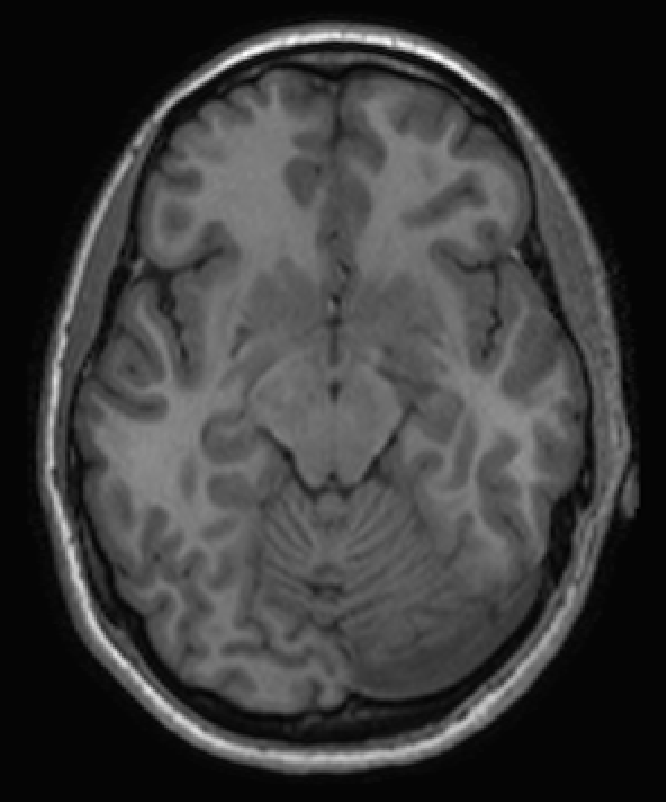
Motivation: PET and MRI images show structural similarity due to the same anatomy.
Acknowledgements: The software uses L-BFGS-B to solve the constrained minimization problem which needs to be downloaded separately. The Fortan code can be obtained from [link, accessed September 2014] and the Matlab interface from [link, accessed September 2014] or [link].







Vector-Valued Image Processing by Parallel Level Sets
Paper: [IEEE Xplore] [preprint (11 MB)]
Download software: [14 MB]
Tested Configurations: The software was tested on Ubuntu 12.04 LTS with Matlab 2013a.
Reference: M. J. Ehrhardt and S. R. Arridge, Vector-Valued Image Processing by Parallel Level Sets , IEEE Transactions on Image Processing, Volume 23, Number 1, Pages 9-18, 2014
Idea: By penalizing parallel level sets two images evolve to a common shape.



Acknowledgements: The original images are part of the training data set of the Berkeley Segmentation Dataset, [download (21 MB), accessed March 2013]. The images have different names in the data base: leopard = 159029, lake = 176035, pyramid = 299091, bugs = 35008, wolf = 42078. The noisy images can be found on Francisco J. Estrada's web page, [download (1 GB), accessed March 2013]. The images have different names on this page: leopard = 0072, lake = 0094, pyramid = 0194, bugs = 0213, wolf = 0246.
We used the Structural Similarity as a figure of merit. To make use of it in the software please download the file ssim_index.m [download (6 KB), accessed March 2013] available from the author's web page.
